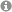OrderedList
- Similarities of ordered gene lists
An R/Bioconductor package to explore the similarity between two lists of ordered genes.
OrderedList
![]()
The methods of the R/Bioconductor compliant package OrderedList provide a comparison of comparisons. Say, we compare two gene expression studies. Both are comparisons of two conditions. Preferably, one condition relates to a good outcome or prognosis and the other one relates to a bad outcome. For each study separately, we might conduct a two-sample test per gene to discover differentially expressed genes. Although each single study might not necessarily reveal significant changes, we may observe considerable overlap in the top-ranking genes. Hence, we wish to compare the results of the two comparisons.
We assign a similarity score to a comparison of two ranked (ordered) gene lists. The similarity score is based on the number of overlapping genes in the top ranks. For each rank, the size of overlap is computed. The final score is in principle a weighted sum of these values, with more weight put on the top ranks.
| Figure: Displayed are the numbers of overlapping genes in the comparison of two gene lists. The overlap size is drawn as a step function over the respective ranks. Top ranks correspond to up-regulated and bottom ranks to down-regulated genes. In addition, the expected overlap and 95% confidence intervals derived from a hypergeometric distribution are shown. |
Publications
- OrderedList - a Bioconductor Package for DetectingSimilarity in Ordered Gene Lists
Lottaz C, Yang X, Scheid S and Spang R.
Bioinformatics, 2006, 22 (18): 2315-2316.
[?pmid: 16844712?|? doi: 10.1093/bioinformatics/btl385 ] - Similarities of ordered gene lists
Yang X, Bentink S, Scheid S, and Spang R.
Journal of Bioinformatics and Computational Biology., 2006, 4 (3): 693-708.
[?pmid: 16960970?|? doi : 10.1142/S0219720006002120?] - Similarities of Ordered Gene Lists - User's Guide to the Bioconductor Package OrderedList
Scheid S, Lottaz C, Yang X, and Spang R.
CompDiag Technical Report Nr. 2006/01 Mar. 2006
Download
Download the R package directly from the Bioconductor WWW-site. To run the package, you need R and the Bioconductor packages Biobase and twilight. Package OrderedList includes a user's manual which can be downloaded separately from above.